Statues of 2025









I’m running out of time for a 2025 post, and thought I’d share some statues of the year. You can try to guess the locations, I’ll put the solution at the bottom.
I attended my first UCR graduation – for my former student Xianghao Kong who is now at an AI startup called Voia. I went to a few conferences this year, including NeurIPS where I had a lot of fun conversations but forgot to take any pictures.
For research, I started a new direction in 2025 and I hope we get to show some results from this in 2026. LLMs and diffusion models generate “information” in some fixed path (one word at a time, or denoise all pixels by a fixed amount each step). Is it possible to discover your own path?
Personally, I started dabbling with chess again after not playing for many years. I’m not sure I have enough spare brain cells to get in good form, but I’ve enjoyed following the drama in the world chess scene – this must be what it is like to be a sports fan.
Solutions: UCR bear is in the middle of our campus. There are two from the Allerton conference at UIUC, the mammoth and Lincoln. The bearded waving guy is in Laguna beach – they have a history of this explained by the plaque (and an actual person who carries on the tradition by waving to everybody). The Kiss of Death is in a cemetery in Barcelona. The newspaper statue is in Culver City, I’m not sure the story, I was just biding time before a nice dinner with former students and colleagues. Godzilla is from Tokyo of course. The log is a “caga tio”, a delightful Christmas tradition from the Catalans in Barcelona. The guy with a bird on his shoulder is from a restaurant near Los Alamos, I’m fondly remembering a fun dinner there.
Filed under: Posted by Greg Ver Steeg | Leave a Comment
NeurIPS 2024
 I’m attending NeurIPS 2024, and within five seconds of walking in the door, there’s a happy lab reunion! Left is Rob Brekelmans. After finishing his PhD with me and Aram at USC, he took a prestigious postdoc fellowship at Vector Institute. After his 2024 ICML best paper award, you better believe he’ll get snapped up on the faculty hiring marking, so better move quickly if you’re hiring! On the right side is Sami Abu-el-Haija, a wizard with graph neural networks, who is now working at Google Research.
I’m attending NeurIPS 2024, and within five seconds of walking in the door, there’s a happy lab reunion! Left is Rob Brekelmans. After finishing his PhD with me and Aram at USC, he took a prestigious postdoc fellowship at Vector Institute. After his 2024 ICML best paper award, you better believe he’ll get snapped up on the faculty hiring marking, so better move quickly if you’re hiring! On the right side is Sami Abu-el-Haija, a wizard with graph neural networks, who is now working at Google Research.
Let’s get back to the reason I’m here. Here’s a nice video from the lead author Yunshu Wu: (though I think to see the video you may have to be registered for the conference). Unfortunately she couldn’t make it to the conference so I’m presenting in her place. It’s a fun and innovative paper making connections between diffusion models, log likelihood ratios, and (noise) classification.
The line of work being exploited here was developed in 2 papers (ICLR 2023, ICLR 2024) by Xianghao Kong. He just successfully defended his thesis proposal, so will also be on the market in June. He’s already done some great industrial research on diffusion models too, at Sony and Adobe, so I’m sure he’ll be in high demand.
Finally, to round out the lab news, I’d like to point out a really cool preprint by a new student, Shaorong Zhang, on diffusion bridge models. And I’m delighted to announce that two more of my USC students: Neal Lawton and Kyle Reing, successfully defended their theses. Congrats!
For Google search, I finally got a new professional portrait, thanks to the UCR photographer Stan Lim. He also does the photography for the prison BA program that my wife helped to start, UCR Lifted, so if you are interested in prison education, you must check out the cool shots there.

Greg Ver Steeg
Associate Professor
CSE
(UCR/Stan Lim)
Filed under: Posted by Greg Ver Steeg | Leave a Comment
One more before ’24
I want to maintain my amazing once-per-year blogging streak, but I spent all my holiday break time on an a new but still secret project that won’t be unveiled until next year 😦
There were lots of exciting developments in 2023, most of which I’m sure I’ll miss.
- Information theoretic diffusion: this approach was introduced in ICLR 2023 and we explore some applications in an ICLR 2024 submission. I feel this perspective still has unique insights to offer (compared to the SDE/ODE/Score matching/VAE/Nonequilibrium thermodynamics perspectives on diffusion). The ICLR 2024 submission was partially inspired by a fun workshop with the small, but fantastic “information decomposition” community, DeMICS.
- It’s been exciting working on more causality stuff again with the smart and prolific Myrl Marmarelis.
- I’ve also spent more time on my role as an Amazon visiting academic. There have been two exciting research developments there that I can’t share until there is a public paper out.
- A few random (but not that personal) developments. (1) I made a cold tub to up the game on my Wim Hof practice (2) doubled my consecutive push-up number (3) I really like the new NYT Connections game.

Filed under: Posted by Greg Ver Steeg | Leave a Comment
UC in 2023
I am happy to announce that I am starting 2023 as an associate professor at the University of California, Riverside, in the computer science department. While I will miss USC, I hope to maintain close connections with the many wonderful colleagues and students I have had the privilege of working with there (I will continue to have a title as an adjunct research associate professor at USC). I will also continue my role as a part-time visiting academic at Amazon Alexa AI.
I am looking forward to using the freedom of tenure to explore several exciting new research directions, see here for a preview of one new direction. I was fortunate to participate in many exciting projects in 2022, here is a sampling of some major developments.
- My PhD student Rob Brekelmans defended and is now a postdoc at Vector Institute. His ICLR 2022 paper presented exciting new results on mutual information estimation (talk).
- A second PhD student in my group defended. Sami Abu-al-Haija is now at Google Research. He contributed a number of significant and influential ideas in graph representation learning.
- Umang Gupta will defend in 2023. His work at ACL 2022 on reducing bias in text generation seems especially relevant given the recent interest in ChatGPT.
- Hrayr Harutyunyan will also defend in 2023. He has made some major contributions on the deep problem of understanding generalization in neural networks, building on his already influential NeurIPS 2021 paper with recent results in ITW in 2022.
- Material science! This is my first paper in material science, led by the multi-talented physicist Marcin Abram. A paper appeared in npj Computational Materials, showing an intriguing way to use self-supervised machine learning to discover changes in material microstructure.
- Neuroimaging. Several papers build on our ongoing work on harmonizing MRIs across sites, and on federated learning. An older line of work of mine, CorEx, saw some new applications in neuroimaging with my first cover article for a journal, an issue of Entropy (shown below).


Filed under: Posted by Greg Ver Steeg | Leave a Comment
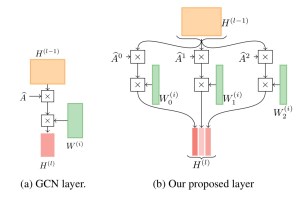
Accelerating graph learning
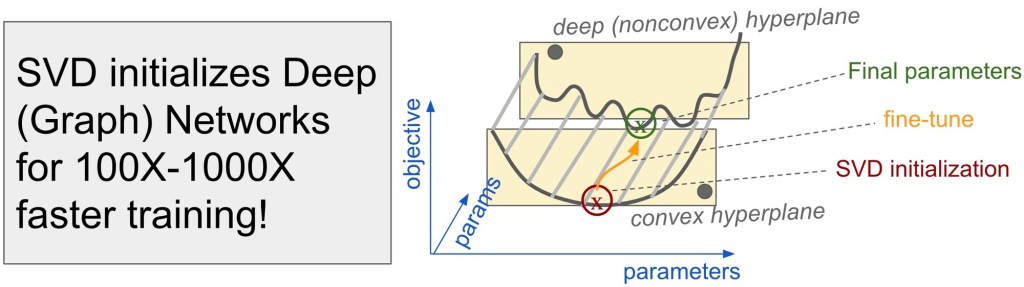
There’s lots of exciting work recently that I haven’t had time to describe. Sami wrote a nice blog post about his NeurIPS 2021 paper on dramatically speeding up graph representation learning with an implicit form of SVD.
I will continue to be too busy to blog much, especially because of the new class I’m teaching on dynamics of representation learning.
Filed under: Posted by Greg Ver Steeg | Leave a Comment
Fairly Accurate Machine Learning
A few posts back, I talked about how fairness could be related to information theory. By removing any information that could be used to identify a group, you make it impossible to give that group preferential treatment. A talented student in our group, Umang Gupta, has taken that line of reasoning further and shown how information theory can give guarantees about trade-offs between fairness and accuracy for some task. Umang made this cool 1 minute explainer video for his paper which will appear in AAAI that sums it up better than I can.
Filed under: Posted by Greg Ver Steeg | Leave a Comment
I’m excited to share a student paper that was just accepted to ICML. Neural networks are capable of memorizing training labels, but if this happens they will generalize poorly when applied to test data. Where is that information about memorized labels stored? Well, it has to be stored in the neural network weights somewhere. You can write down an information measure that captures the amount of memorization. Unfortunately, it’s very difficult to estimate or control this term, because it involves high-dimensional quantities. Hrayr found an interesting way to control this information, by using a separate neural network that estimates gradients without relying too much on label information.

One of the most fun parts of this project is that we can then check which labels in a dataset seem to require the most memorization. In the figure, you can see that the labels provided by humans (y) for this dataset are often wrong or confusing. Our network that tries to learn a classifier without memorizing labels often does better (y hat).
Filed under: Posted by Greg Ver Steeg | Leave a Comment
ICML and MixHop
ICML 2019 is coming up soon, and I plan to be there (except I’m missing Tuesday). I want to briefly tout the excellent work of a fantastic student who joined our lab, Sami Abu-El-Haija. If you’ve kept up with develops on learning with graphs, you may be aware of graph convolutional networks, which combine the best of neural networks and spectral learning on graphs to produce some top-notch results on graph datasets. There is one drawback of the original graph convolution approach. Unlike visual convolutions, it doesn’t allow for complex weighting of pixels at different locations within the kernel. Basically, the equivalent relative weighting in graph convolutions are constant, and this makes it difficult to distinguish how neighbors in a graph might systematically differ in their effect from neighbors of neighbors. Sami’s paper MixHop rectifies this issue and shows that this gives some nice performance boosts. Sami will be presenting the work at ICML and code for the approach is also available on github.
Filed under: Posted by Greg Ver Steeg | Leave a Comment
Fairness and Information

Southern information theorists after the civil war realized that although they could no longer exclude former slaves from the polls, they could exclude people based on other criteria like, say, education, and that these criteria happen to be highly correlated with formerly being a slave who was not allowed education. Republican information theorists continue to exploit this observation in ugly ways to exclude voters, like disenfranchising voters without street addresses (many Native Americans).
In the era of big data, these deplorable types of discrimination have become more insidious. Algorithms determine things like what you see on your Facebook feed or whether you are approved for a home loan. Although a loan approval can’t be explicitly based on race, it might depend on your zip code which may be highly correlated for historical reasons.
This leads to an interesting mathematical question: can we design algorithms that are good at predicting things like whether you are a promising applicant for a home loan without being discriminatory? This type of question is the heart of the emerging field of “fair representation learning”.
This is effectively an information theory question. We want to know if our data contains information about the thing that we would like to predict, but that is not informative about some protected variable. The contribution of a great PhD student in my group, Daniel Moyer, to the growing field of fair representation learning was to come up with an explicit and direct information-theoretic characterization of this problem. His results will appear as a paper at NIPS this year.
He showed that an information-theoretic approach could be more effective with less effort than previous approaches which rely on an adversary to test whether any protected information has leaked through. He also showed that you can use this approach in other fun ways. For instance, you can imagine counter-factual answers like what the data would look like if we changed just the protected variable and nothing else. As a concrete visual example, you can imagine that our “protected variable” is the digit in a handwritten picture. Now our neural net learns to represent the image, but without knowing which specific digit was written. Then we can run the neural net in reverse to reconstruct an image that looks similar to the original stylistically but with any value of the digit that we choose.

Fig. 3 from our paper.
Going back to the original motivation about fairness, even though we can define it in this information-theoretic way it’s not clear that this fits a human conception of fairness in all scenarios. Formulating fairness in a way that meets societal goals for different situations and is quantifiable is an ongoing problem in this interesting new field.
Filed under: Posted by Greg Ver Steeg | 1 Comment
No blog-iversary update
It’s officially been a year since my last blog. There have been so many exciting new things going on that it’s been hard to take time out for some nice big picture blog posts. Here are a few areas that I have the best of intentions for getting to.
- Fair representation learning using information theory (2018 NIPS paper)
- Ryan Gallagher’s nice work on anchored topic models.
- Finding coordinated activity in collective sensing games and other spatiotemporal data using structured latent factor discovery. (This is mostly for showing off cool movies.)
- Echo noise. This is a new approach for bounding the information capacity in learning, and allows us to do many cool things like information bottleneck, VAEs, and sparse regression.
- A blessing of dimensionality and how we can use it for better causal inference.
- I’ve still never blogged about our work on neuroimaging or Alzheimer’s disease (see an article here)
- Sahil Garg’s paper for AAAI 2019 on biomedical relation extraction
- Neal Lawton’s work at UAI 2018 and ongoing mission to improve inference in hidden variable models
- Automating data science! (preview here)
Filed under: Posted by Greg Ver Steeg | Leave a Comment